Do you want to use study protein-DNA interaction?
Our supplier Epigentek provides Chromatin Immunoprecipitation Sequencing (ChIP-Seq) Service that is very useful for your research.
ChIP can be used to determine whether a specific protein binds to a particular sequence of a gene, such as the target sequence of a transcription factor or to compare the levels of histone methylation associated with a specific gene promoter region between normal and diseased tissues. Identifying the genetic targets of DNA binding proteins and revealing the mechanism of protein-DNA interaction is crucial for understanding cellular processes.
Map genome-wide histone modifications with EpiGentek’s comprehensive chromatin immunoprecipitation sequencing (ChIP-Seq) platform. Optimized buffers and protocol allow minimal ChIP background and increased sensitivity and specificity of the ChIP reaction.
Highlights
- Map any H2, H3, H4 histone modifications across the entire genome
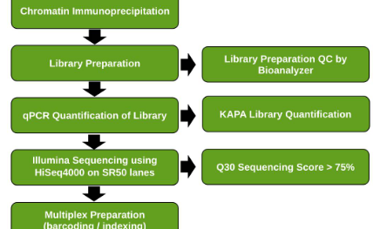
- All kits used in our ChIP-Seq services are commercially available with our ChIP-Seq Workflow.
- Highly efficient enrichment of targeted DNA. Enrichment ratio of positive to negative control > 500.
- High sensitivity and flexibility allows both non-barcoded (singleplexed) and barcoded (multiplexed) DNA library preparation.
- Flexible sample requirements
Delivery of high-quality publishable data (Illumina FASTQ raw sequencing files, FastQC quality control insight, Bismark read mapping, Peak calling, peak annotation and differential binding analysis).